Why use a Cluster?
Overview
Teaching: 15 min
Exercises: 5 minQuestions
Why would I be interested in High Performance Computing (HPC)?
What can I expect to learn from this course?
Objectives
Be able to describe what an HPC system is
Identify how an HPC system could benefit you.
Frequently, research problems that use computing can outgrow the capabilities of the desktop or laptop computer where they started:
-
A statistics student wants to cross-validate a model. This involves running the model 1000 times – but each run takes an hour. Running the model on a laptop will take over a month!
-
A genomics researcher has been using small datasets of sequence data, but soon will be receiving a new type of sequencing data that is 10 times as large. It’s already challenging to open the datasets on a computer – analyzing these larger datasets will probably crash it.
-
An engineer is using a fluid dynamics package that has an option to run in parallel. So far, this option was not utilized on a desktop. In going from 2D to 3D simulations, the simulation time has more than tripled. It might be useful to take advantage of that option or feature.
In all these cases, access to more computers is needed. Those computers should be usable at the same time.
And what do you do?
Talk to your neighbour, office mate or rubber duck about your research.
- How does computing help you do your research?
- How could more computing help you do more or better research?
A standard Laptop for standard tasks
Today, people coding or analysing data typically work with laptops.
Let’s dissect what resources programs running on a laptop require:
- the keyboard and/or touchpad is used to tell the computer what to do (Input)
- the internal computing resources Central Processing Unit and Memory perform calculation
- the display depicts progress and results (Output)
Schematically, this can be reduced to the following:
When tasks take too long
When the task to solve becomes heavy on computations, the operations are typically out-sourced from the local laptop or desktop to elsewhere. Take for example the task to find the directions for your next vacation. The capabilities of your laptop are typically not enough to calculate that route spontaneously: finding the shortest path through a network runs on the order of (v log v) time, where v (vertices) represents the number of intersections in your map. Instead of doing this yourself, you use a website, which in turn runs on a server, that is almost definitely not in the same room as you are.
Note here, that a server is mostly a noisy computer mounted into a rack cabinet which in turn resides in a data center. The internet made it possible that these data centers do not require to be nearby your laptop. What people call the cloud is mostly a web-service where you can rent such servers by providing your credit card details and requesting remote resources that satisfy your requirements. This is often handled through an online, browser-based interface listing the various machines available and their capacities in terms of processing power, memory, and storage.
The server itself has no direct display or input methods attached to it. But most importantly, it has much more storage, memory and compute capacity than your laptop will ever have. In any case, you need a local device (laptop, workstation, mobile phone or tablet) to interact with this remote machine, which people typically call ‘a server’.
When one server is not enough
If the computational task or analysis to complete is daunting for a single server, larger agglomerations of servers are used. These go by the name of “clusters” or “super computers”.
The methodology of providing the input data, configuring the program options, and retrieving the results is quite different to using a plain laptop. Moreover, using a graphical interface is often discarded in favor of using the command line. This imposes a double paradigm shift for prospective users asked to
- work with the command line interface (CLI), rather than a graphical user interface (GUI)
- work with a distributed set of computers (called nodes) rather than the machine attached to their keyboard & mouse
I’ve never used a server, have I?
Take a minute and think about which of your daily interactions with a computer may require a remote server or even cluster to provide you with results.
Some Ideas
- Checking email: your computer (possibly in your pocket) contacts a remote machine, authenticates, and downloads a list of new messages; it also uploads changes to message status, such as whether you read, marked as junk, or deleted the message. Since yours is not the only account, the mail server is probably one of many in a data center.
- Searching for a phrase online involves comparing your search term against a massive database of all known sites, looking for matches. This “query” operation can be straightforward, but building that database is a monumental task! Servers are involved at every step.
- Searching for directions on a mapping website involves connecting your (A) starting and (B) end points by traversing a graph in search of the “shortest” path by distance, time, expense, or another metric. Converting a map into the right form is relatively simple, but calculating all the possible routes between A and B is expensive.
Checking email could be serial: your machine connects to one server and exchanges data. Searching by querying the database for your search term (or endpoints) could also be serial, in that one machine receives your query and returns the result. However, assembling and storing the full database is far beyond the capability of any one machine. Therefore, these functions are served in parallel by a large, “hyperscale” collection of servers working together.
Key Points
High Performance Computing (HPC) typically involves connecting to very large computing systems elsewhere in the world.
These other systems can be used to do work that would either be impossible or much slower on smaller systems.
The standard method of interacting with such systems is via a command line interface called Bash.
Working on a remote HPC system
Overview
Teaching: 25 min
Exercises: 10 minQuestions
What is an HPC system?
How does an HPC system work?
How do I log on to a remote HPC system?
Objectives
Connect to a remote HPC system.
Understand the general HPC system architecture.
What is an HPC system?
The words “cloud”, “cluster”, and the phrase “high-performance computing” or “HPC” are used a lot in different contexts and with various related meanings. So what do they mean? And more importantly, how do we use them in our work?
The cloud is a generic term commonly used to refer to computing resources that are a) provisioned to users on demand or as needed and b) represent real or virtual resources that may be located anywhere on Earth. For example, a large company with computing resources in Brazil, Zimbabwe and Japan may manage those resources as its own internal cloud and that same company may also utilize commercial cloud resources provided by Amazon or Google. Cloud resources may refer to machines performing relatively simple tasks such as serving websites, providing shared storage, providing webservices (such as e-mail or social media platforms), as well as more traditional compute intensive tasks such as running a simulation.
The term HPC system, on the other hand, describes a stand-alone resource for computationally intensive workloads. They are typically comprised of a multitude of integrated processing and storage elements, designed to handle high volumes of data and/or large numbers of floating-point operations (FLOPS) with the highest possible performance. For example, all of the machines on the Top-500 list are HPC systems. To support these constraints, an HPC resource must exist in a specific, fixed location: networking cables can only stretch so far, and electrical and optical signals can travel only so fast.
The word “cluster” is often used for small to moderate scale HPC resources less impressive than the Top-500. Clusters are often maintained in computing centers that support several such systems, all sharing common networking and storage to support common compute intensive tasks.
Logging in
The first step in using a cluster is to establish a connection from our laptop to the cluster, via the Internet and/or your organisation’s network. We use a program called the Secure SHell (or ssh) client for this. Make sure you have a SSH client installed on your laptop. Refer to the setup section for more details.
Go ahead and log in to the Plato HPC cluster at the University of Saskatchewan.
[user@laptop ~]$ ssh nsid@plato.usask.ca
Remember to replace nsid by your own NSID. You will be asked
for your password; this is the same one you use with other University of Saskatchewan resources, such as the PAWS website. Watch out: the characters you type after the password prompt are not displayed on
the screen. Normal output will resume once you press Enter.
You are logging in using a program known as the secure shell or ssh. This establishes a temporary
encrypted connection between your laptop and plato.usask.ca. The word before the @
symbol, e.g. nsid here, is the user account name that you have permission to use on the
cluster.
Where are we?
Very often, many users are tempted to think of a high-performance computing installation as one
giant, magical machine. Sometimes, people will assume that the computer they’ve logged onto is the
entire computing cluster. So what’s really happening? What computer have we logged on to? The name
of the current computer we are logged onto can be checked with the hostname command. (You may also
notice that the current hostname is also part of our prompt!)
[nsid@platolgn01 ~]$ hostname
platolgn01
What’s in your home directory?
The system administrators may have configured your home directory with some helpful files, folders, and links (shortcuts) to space reserved for you on other filesystems. Take a look around and see what you can find.
Home directory contents vary from user to user. Please discuss any differences you spot with your neighbors.
Hint: The shell commands
pwdandlsmay come in handy.Solution
Use
pwdto print the working directory path:[nsid@platolgn01 ~]$ pwdThe deepest layer should differ: nsid is uniquely yours. Are there differences in the path at higher levels?
You can run
lsto list the directory contents, though it’s possible nothing will show up (if no files have been provided). To be sure, use the-aflag to show hidden files, too.[nsid@platolgn01 ~]$ ls -aAt a minimum, this will show the current directory as
., and the parent directory as...If both of you have empty directories, they will look identical. If you or your neighbor has used the system before, there may be differences. What are you working on?
Nodes
Individual computers that compose a cluster are typically called nodes (although you will also hear people call them servers, computers and machines). On a cluster, there are different types of nodes for different types of tasks. The node where you are right now is called the login node or submit node. A login node serves as an access point to the cluster. As a gateway, it is well suited for uploading and downloading files, setting up software, and running quick tests. It should never be used for doing computationally intensive work.
The real work on a cluster gets done by the worker (or compute) nodes. Worker nodes come in many shapes and sizes, but generally are dedicated to long or hard tasks that require a lot of computational resources.
All interaction with the worker nodes is handled by a specialized piece of software called a scheduler (the scheduler used in this lesson is called ). We’ll learn more about how to use the scheduler to submit jobs next, but for now, it can also tell us more information about the worker nodes.
For example, we can view all of the worker nodes with the sinfo command.
[nsid@platolgn01 ~]$ sinfo
PARTITION AVAIL TIMELIMIT NODES STATE NODELIST
plato_long up 21-00:00:0 25 mix plato[417-421,423,425-426,428-435,437-441,445-448]
plato_long up 21-00:00:0 62 idle plato[225-234,237-238,240-247,309-330,333-337,339-343,346-348,422,424,427,436,442-444]
plato_medium up 4-00:00:00 25 mix plato[417-421,423,425-426,428-435,437-441,445-448]
plato_medium up 4-00:00:00 62 idle plato[225-234,237-238,240-247,309-330,333-337,339-343,346-348,422,424,427,436,442-444]
plato_short up 12:00:00 25 mix plato[417-421,423,425-426,428-435,437-441,445-448]
plato_short up 12:00:00 62 idle plato[225-234,237-238,240-247,309-330,333-337,339-343,346-348,422,424,427,436,442-444]
plato_gpu up 7-00:00:00 4 idle plato-base-gpu-[23003-23004],platogpu[103-104]
plato_gpu_short up 12:00:00 4 idle plato-base-gpu-[23003-23004],platogpu[103-104]
[...]
There are also specialized machines used for managing disk storage, user authentication, and other infrastructure-related tasks. Although we do not typically logon to or interact with these machines directly, they enable a number of key features like ensuring our user account and files are available throughout the HPC system.
What’s in a node?
All of the nodes in an HPC system have the same components as your own laptop or desktop: CPUs (sometimes also called processors or cores), memory (or RAM), and disk space. CPUs are a computer’s tool for actually running programs and calculations. Information about a current task is stored in the computer’s memory. Disk refers to all storage that can be accessed like a file system. This is generally storage that can hold data permanently, i.e. data is still there even if the computer has been restarted. While this storage can be local (a hard drive installed inside of it), it is more common for nodes to connect to a shared, remote fileserver or cluster of servers.
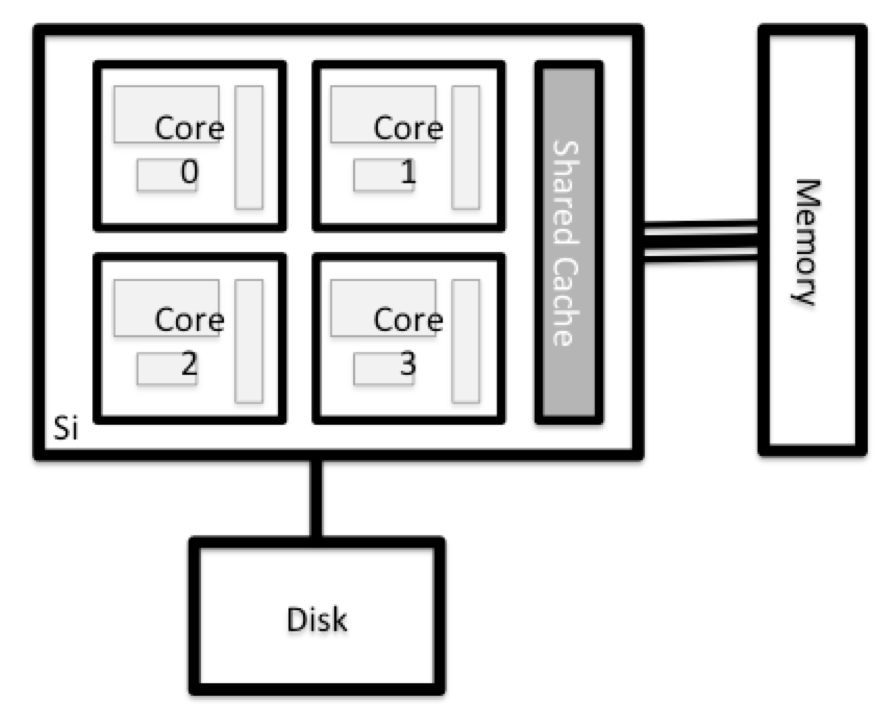
Explore Your Computer
Try to find out the number of CPUs and amount of memory available on your personal computer.
Note that, if you’re logged in to the remote computer cluster, you need to log out first. To do so, type
Ctrl+dorexit:[nsid@platolgn01 ~]$ exit [user@laptop ~]$Solution
There are several ways to do this. Most operating systems have a graphical system monitor, like the Windows Task Manager. More detailed information can be found on the command line:
- Run system utilities
[user@laptop ~]$ nproc --all [user@laptop ~]$ free -m- Read from
/proc[user@laptop ~]$ cat /proc/cpuinfo [user@laptop ~]$ cat /proc/meminfo- Run system monitor
[user@laptop ~]$ htop
Explore The Head Node
Now compare the resources of your computer with those of the head node.
Solution
[user@laptop ~]$ ssh nsid@plato.usask.ca [nsid@platolgn01 ~]$ nproc --all [nsid@platolgn01 ~]$ free -mYou can get more information about the processors using
lscpu, and a lot of detail about the memory by reading the file/proc/meminfo:[nsid@platolgn01 ~]$ less /proc/meminfoYou can also explore the available filesystems using
dfto show disk free space. The-hflag renders the sizes in a human-friendly format, i.e., GB instead of B. The type flag-Tshows what kind of filesystem each resource is.[nsid@platolgn01 ~]$ df -ThThe local filesystems (ext, tmp, xfs, zfs) will depend on whether you’re on the same login node (or compute node, later on). Networked filesystems (beegfs, cifs, gpfs, nfs, pvfs) will be similar – but may include yourUserName, depending on how it is mounted.
Shared file systems
This is an important point to remember: files saved on one node (computer) are often available everywhere on the cluster!
Explore a Worker Node
Finally, let’s look at the resources available on the worker nodes where your jobs will actually run. Try running this command to see the name, CPUs and memory available on the worker nodes:
[nsid@platolgn01 ~]$ sinfo -n plato418 -o "%n %c %m"
Compare Your Computer, the Head Node and the Worker Node
Compare your laptop’s number of processors and memory with the numbers you see on the cluster head node and worker node. Discuss the differences with your neighbor.
What implications do you think the differences might have on running your research work on the different systems and nodes?
Differences Between Nodes
Many HPC clusters have a variety of nodes optimized for particular workloads. Some nodes may have larger amount of memory, or specialized resources such as Graphical Processing Units (GPUs).
With all of this in mind, we will now cover how to talk to the cluster’s scheduler, and use it to start running our scripts and programs!
Key Points
An HPC system is a set of networked machines.
HPC systems typically provide login nodes and a set of worker nodes.
The resources found on independent (worker) nodes can vary in volume and type (amount of RAM, processor architecture, availability of network mounted file systems, etc.).
Files saved on one node are available on all nodes.
Scheduling jobs
Overview
Teaching: 45 min
Exercises: 30 minQuestions
What is a scheduler and why are they used?
How do I launch a program to run on any one node in the cluster?
How do I capture the output of a program that is run on a node in the cluster?
Objectives
Run a simple Hello World style program on the cluster.
Submit a simple Hello World style script to the cluster.
Use the batch system command line tools to monitor the execution of your job.
Inspect the output and error files of your jobs.
Job scheduler
An HPC system might have thousands of nodes and thousands of users. How do we decide who gets what and when? How do we ensure that a task is run with the resources it needs? This job is handled by a special piece of software called the scheduler. On an HPC system, the scheduler manages which jobs run where and when.
The following illustration compares these tasks of a job scheduler to a waiter in a restaurant. If you can relate to an instance where you had to wait for a while in a queue to get in to a popular restaurant, then you may now understand why sometimes your job do not start instantly as in your laptop.
The scheduler used in this lesson is SLURM. Although SLURM is not used everywhere, running jobs is quite similar regardless of what software is being used. The exact syntax might change, but the concepts remain the same.
Running a batch job
The most basic use of the scheduler is to run a command non-interactively. Any command (or series of commands) that you want to run on the cluster is called a job, and the process of using a scheduler to run the job is called batch job submission.
In this case, the job we want to run is just a shell script. Let’s create a demo shell script to
run as a test. The cluster offers a number of terminal-based text editors. Use
whichever you prefer. Unsure? nano is a pretty good, basic choice.
[nsid@platolgn01 ~]$ nano example-job.sh
[nsid@platolgn01 ~]$ chmod +x example-job.sh
[nsid@platolgn01 ~]$ cat example-job.sh
#!/bin/bash
echo -n "This script is running on "
hostname
sleep 20
echo "Job done!"
Creating our test job
Run the script. Does it execute on the cluster or just our login node?
Solution
[nsid@platolgn01 ~]$ ./example-job.shThis script is running on platolgn01 Job done!This job runs on the login node.
If you completed the previous challenge successfully, you probably realise that there is a
distinction between running the job through the scheduler and just “running it”. To submit this job
to the scheduler, we use the sbatch command.
[nsid@platolgn01 ~]$ sbatch example-job.sh
Submitted batch job 736234
And that’s all we need to do to submit a job. Our work is done – now the scheduler takes over and
tries to run the job for us. While the job is waiting to run, it goes into a list of jobs called
the queue. To check on our job’s status, we check the queue using the command
squeue -u nsid.
[nsid@platolgn01 ~]$ squeue -u nsid
JOBID USER ACCOUNT NAME ST TIME_LEFT NODES CPUS GRES MIN_MEM NODELIST (REASON)
726557 nsid hpc_s_worksh example-job.sh R 19:50 1 1 (null) 512M plato418 (None)
We can see all the details of our job, most importantly that it is in the R or RUNNING state.
Sometimes our jobs might need to wait in a queue (PD, for pending) or have an error (E).
Once a job is finished, it
will no longer be listed by squeue.
Where’s the output?
On the login node, this script printed output to the terminal – but when our job finishes, there’s nothing. Where’d it go?
Cluster job output is typically redirected to a file in the directory you launched it from. Use
lsto find the file andcatto read it.
Customising a job
The job we just ran used all of the scheduler’s default options. In a real-world scenario, that’s probably not what we want. The default options represent a reasonable minimum. Chances are, we will need more cores, more memory, more time, among other special considerations. To get access to these resources we must customize our job script.
Comments in UNIX shell scripts (denoted by #) are typically ignored, but there are exceptions.
For instance the special #! comment at the beginning of scripts specifies what program should be
used to run it (you’ll typically see #!/bin/bash). Schedulers like SLURM also
have a special comment used to denote special scheduler-specific options. Though these comments
differ from scheduler to scheduler, SLURM’s special comment is
#SBATCH. Anything following the #SBATCH comment is interpreted
as an instruction to the scheduler.
Let’s illustrate this by example. By default, a job’s name is the name of the script, but the
-J option can be used to change the name of a job. Add an option to the
script:
[nsid@platolgn01 ~]$ cat example-job.sh
#!/bin/bash
#SBATCH -J new_name
echo -n "This script is running on "
hostname
sleep 20
echo "Job done!"
Submit the job (using sbatch example-job.sh)
and monitor it:
[nsid@platolgn01 ~]$ squeue -u nsid
JOBID USER ACCOUNT NAME ST TIME_LEFT NODES CPUS GRES MIN_MEM NODELIST (REASON)
726970 nsid hpc_s_worksh new_name R 19:55 1 1 (null) 512M plato418 (None)
Fantastic, we’ve successfully changed the name of our job!
Setting up email notifications
Jobs on an HPC system might run for days or even weeks. We probably have better things to do than constantly check on the status of our job with
squeue. Looking at the manual page forsbatch, can you set up our test job to send you an email when it finishes?Hint
You can use the manual pages for SLURM utilities to find more about their capabilities. On the command line, these are accessed through the
manutility: runman <program-name>. You can find the same information online by searching “man". [nsid@platolgn01 ~]$ man sbatch
Resource requests
But what about more important changes, such as the number of cores and memory for our jobs? One thing that is absolutely critical when working on an HPC system is specifying the resources required to run a job. This allows the scheduler to find the right time and place to schedule our job. If you do not specify requirements (such as the amount of time you need), you will likely be stuck with your site’s default resources, which is probably not what you want.
The following are several key resource requests:
-
--ntasks=<ntasks>or-n <ntasks>: How many parallel tasks will your job start? Tasks may be started on the same, or on different nodes. -
--time=<days-hours:minutes:seconds>or-t <days-hours:minutes:seconds>: How much real-world time (walltime) will your job take to run? The<days>part can be omitted. -
--mem=<megabytes>: How much memory on a node does your job need in megabytes? You can also specify gigabytes using by adding “G” afterwards (example:--mem=5G) -
--nodes=<nnodes>or-N <nnodes>, and--ntasks-per-node=<ntasks>: How many nodes does your job need to run on and how many tasks will it run on each? Note that you should use either these options or--ntasks, but never combine--nodesand--ntasks.
Note that just requesting these resources does not make your job run faster! We’ll talk more about how to make sure that you’re using resources effectively in a later episode of this lesson.
Submitting resource requests
Submit a job that requests 1 full node and 1 minute of walltime.
Solution
[nsid@platolgn01 ~]$ cat example-job.sh#!/bin/bash #SBATCH --time=00:01:00 #SBATCH --nodes=1 #SBATCH --ntasks-per-node=40 #SBATCH --mem=185G echo -n "This script is running on " hostname sleep 20 # time in seconds echo "This script has finished successfully."[nsid@platolgn01 ~]$ sbatch example-job.sh
Job environment variables
When SLURM runs a job, it sets a number of environment variables for the job. One of these will let us check what directory our job script was submitted from. The
SLURM_SUBMIT_DIRvariable is set to the directory from which our job was submitted. Using theSLURM_SUBMIT_DIRvariable, modify your job so that it prints out the location from which the job was submitted.Solution
[nsid@platolgn01 ~]$ nano example-job.sh [nsid@platolgn01 ~]$ cat example-job.sh#!/bin/bash #SBATCH -t 00:00:30 echo -n "This script is running on " hostname echo "This job was launched in the following directory:" echo ${SLURM_SUBMIT_DIR}
Resource requests are typically binding. If you exceed them, your job will be killed. Let’s use walltime as an example. We will request 1 minute of walltime, and attempt to run a job for two minutes.
[nsid@platolgn01 ~]$ cat example-job.sh
#!/bin/bash
#SBATCH -J long_job
#SBATCH -t 00:01:00
echo -n "This script is running on "
hostname
sleep 120 # time in seconds
echo "This script has finished successfully."
Submit the job and wait for it to finish. Once it is has finished, check the log file.
[nsid@platolgn01 ~]$ sbatch example-job.sh
[nsid@platolgn01 ~]$ squeue -u nsid
[nsid@platolgn01 ~]$ cat slurm-38193.out
This job is running on:
plato418
slurmstepd: error: *** JOB 38193 ON plato418 CANCELLED AT 2020-10-06T16:35:48 DUE TO TIME LIMIT ***
Our job was killed for exceeding the amount of resources it requested. Although this appears harsh, this is actually a feature. Strict adherence to resource requests allows the scheduler to find the best possible place for your jobs. Even more importantly, it ensures that another user cannot use more resources than they’ve been given. If another user messes up and accidentally attempts to use all of the cores or memory on a node, SLURM will either restrain their job to the requested resources or kill the job outright. Other jobs on the node will be unaffected. This means that one user cannot mess up the experience of others, the only jobs affected by a mistake in scheduling will be their own.
Cancelling a job
Sometimes we’ll make a mistake and need to cancel a job. This can be done with the
scancel command. Let’s submit a job and then cancel it using its job number (remember
to change the walltime so that it runs long enough for you to cancel it before it is killed!).
[nsid@platolgn01 ~]$ sbatch example-job.sh
[nsid@platolgn01 ~]$ squeue -u nsid
Submitted batch job 726557
JOBID USER ACCOUNT NAME ST TIME_LEFT NODES CPUS GRES MIN_MEM NODELIST (REASON)
726557 nsid hpc_s_worksh example-job.sh R 19:50 1 1 (null) 512M plato418 (None)
Now cancel the job with its job number (printed in your terminal). A clean return of your command prompt indicates that the request to cancel the job was successful.
[nsid@platolgn01 ~]$ scancel 726557
# ... Note that it might take a minute for the job to disappear from the queue ...
[nsid@platolgn01 ~]$ squeue -u nsid
JOBID USER ACCOUNT NAME ST TIME_LEFT NODES CPUS GRES MIN_MEM NODELIST (REASON)
Cancelling multiple jobs
We can also cancel all of our jobs at once using the
-uoption. This will delete all jobs for a specific user (in this case, yourself). Note that you can only delete your own jobs.Try submitting multiple jobs and then cancelling them all.
Solution
First, submit a trio of jobs:
[nsid@platolgn01 ~]$ sbatch example-job.sh [nsid@platolgn01 ~]$ sbatch example-job.sh [nsid@platolgn01 ~]$ sbatch example-job.shThen, cancel them all:
[nsid@platolgn01 ~]$ scancel -u nsid
Other types of jobs
Up to this point, we’ve focused on running jobs in batch mode. SLURM also provides the ability to start an interactive session.
There are very frequently tasks that need to be done interactively. Creating an entire job
script might be overkill, but the amount of resources required is too much for a login node to
handle. A good example of this might be building a genome index for alignment with a tool like
HISAT2. Fortunately, we can run these types of
tasks as a one-off with srun.
srun runs a single command on the cluster and then exits. Let’s
demonstrate this by running the hostname command with srun. (We can
cancel an srun job with Ctrl-c.)
[nsid@platolgn01 ~]$ srun hostname
plato418
srun accepts all of the same options as sbatch.
However, instead of specifying these in a script, these options are specified on the command-line
when starting a job. To submit a job that uses 2 CPUs for instance, we could use the following
command:
[nsid@platolgn01 ~]$ srun -n 2 echo "This job will use 2 CPUs."
This job will use 2 CPUs.
This job will use 2 CPUs.
Typically, the resulting shell environment will be the same as that for
sbatch.
Interactive jobs
Sometimes, you will need a lot of resource for interactive use. Perhaps it’s our first time running
an analysis or we are attempting to debug something that went wrong with a previous job.
Fortunately, SLURM makes it easy to start an interactive job with srun:
[nsid@platolgn01 ~]$ salloc
You should be presented with a bash prompt. Note that the prompt will likely change to reflect your
new location, in this case the compute node we are logged on. You can also verify this with
hostname.
Creating remote graphics
To see graphical output inside your jobs, you need to use X11 forwarding. To connect with this feature enabled, use the
-Yoption when you login with thesshcommand, e.g.,ssh -Y nsid@plato.usask.ca.To demonstrate what happens when you create a graphics window on the remote node, use the
xeyescommand. A relatively adorable pair of eyes should pop up (pressCtrl-Cto stop). If you are using a Mac, you must have installed XQuartz (and restarted your computer) for this to work.
When you are done with the interactive job, type exit to quit your session.
Key Points
The scheduler handles how compute resources are shared between users.
Everything you do should be run through the scheduler.
A job is just a shell script.
If in doubt, request more resources than you will need.
Accessing software
Overview
Teaching: 30 min
Exercises: 15 minQuestions
How do we load and unload software packages?
Objectives
Understand how to load and use a software package.
On a high-performance computing system, it is seldom the case that the software we want to use is available when we log in. It is installed, but we will need to “load” it before it can run.
Before we start using individual software packages, however, we should understand the reasoning behind this approach. The three biggest factors are:
- software incompatibilities
- versioning
- dependencies
Software incompatibility is a major headache for programmers. Sometimes the presence (or absence) of
a software package will break others that depend on it. Two of the most famous examples are Python 2
and 3 and C compiler versions. Python 3 famously provides a python command that conflicts with
that provided by Python 2. Software compiled against a newer version of the C libraries and then
used when they are not present will result in a nasty 'GLIBCXX_3.4.20' not found error, for
instance.
Software versioning is another common issue. A team might depend on a certain package version for their research project - if the software version was to change (for instance, if a package was updated), it might affect their results. Having access to multiple software versions allow a set of researchers to prevent software versioning issues from affecting their results.
Dependencies are where a particular software package (or even a particular version) depends on having access to another software package (or even a particular version of another software package). For example, the VASP materials science software may depend on having a particular version of the FFTW (Fastest Fourier Transform in the West) software library available for it to work.
Environment modules
Environment modules are the solution to these problems. A module is a self-contained description of a software package - it contains the settings required to run a software package and, usually, encodes required dependencies on other software packages.
There are a number of different environment module implementations commonly
used on HPC systems: the two most common are TCL modules and Lmod. Both of
these use similar syntax and the concepts are the same so learning to use one will
allow you to use whichever is installed on the system you are using. In both
implementations the module command is used to interact with environment modules. An
additional subcommand is usually added to the command to specify what you want to do. For a list
of subcommands you can use module -h or module help. As for all commands, you can
access the full help on the man pages with man module.
On login you may start out with a default set of modules loaded or you may start out with an empty environment; this depends on the setup of the system you are using.
Listing available modules
To see available software modules, use module avail
[nsid@platolgn01 ~]$ module avail
[Some output removed for clarity]
------------------------------- MPI-dependent avx2 modules -------------------------------
abyss/2.1.5 (bio) neuron/8.0.0 (bio,D)
abyss/2.2.5 (bio,D) opencarp/4.0
adol-c/2.7.2 opencascade/7.5.2 (D)
alpscore/2.2.0 (phys,D) opencoarrays/2.9.2
amber/18.14-18.17 (chem) openfoam-extend/4.1 (phys)
amber/20.9-20.15 (chem) openfoam/v2006 (phys)
amber/20.12-20.15 (chem,D) openfoam/v2012 (phys)
ambertools/20 openfoam/v2112 (phys)
ambertools/21 openfoam/v2206 (phys)
ambertools/23 (D) openfoam/v2212 (phys)
apbs/1.3 (chem) openfoam/v2306 (phys)
arpack-ng/3.9.0 (math,D) openfoam/8 (phys)
aspect/2.3.0 openfoam/9 (phys)
aspect/2.4.0 (D) openfoam/10 (phys)
astrid/2.2.1 openfoam/11 (phys,D)
blacs/1.1 (math) openmc/0.13.2
boost-mpi/1.72.0 (t) openmc/0.13.3 (D)
boost-mpi/1.80.0 (t,D) openmm-alphafold/7.5.1
cantera/2.4.0 (chem) openmm/7.5.0 (chem)
cantera/2.5.1 (chem) openmm/7.7.0 (chem)
cantera/2.6.0 (chem,D) openmm/8.0.0 (chem,D)
cdo/1.9.8 (geo) opensees/3.2.0
cdo/2.0.4 (geo) opensees/3.5.0 (D)
cdo/2.0.5 (geo) orca/4.2.1 (chem)
cdo/2.2.1 (geo,D) p4est/2.2 (math)
cgns/3.4.1 (phys) p4est/2.3.2 (math)
cgns/4.1.0 (phys) p4est/2.8.5 (math,D)
cgns/4.1.2 (phys,D) paraview-offscreen-gpu/5.8.0 (vis)
cp2k/8.2 (chem) paraview-offscreen-gpu/5.9.1 (vis)
[Most output removed for clarity]
Where:
S: Module is Sticky, requires --force to unload or purge
bio: Bioinformatic libraries/apps / Logiciels de bioinformatique
m: MPI implementations / Implémentations MPI
math: Mathematical libraries / Bibliothèques mathématiques
L: Module is loaded
io: Input/output software / Logiciel d'écriture/lecture
t: Tools for development / Outils de développement
vis: Visualisation software / Logiciels de visualisation
chem: Chemistry libraries/apps / Logiciels de chimie
geo: Geography libraries/apps / Logiciels de géographie
phys: Physics libraries/apps / Logiciels de physique
Aliases: Aliases exist: foo/1.2.3 (1.2) means that "module load foo/1.2" will load foo/1.2.3
D: Default Module
If the avail list is too long consider trying:
"module --default avail" or "ml -d av" to just list the default modules.
"module overview" or "ml ov" to display the number of modules for each name.
Use "module spider" to find all possible modules and extensions.
Use "module keyword key1 key2 ..." to search for all possible modules matching any of the "keys".
Listing currently loaded modules
You can use the module list command to see which modules you currently have loaded
in your environment. If you have no modules loaded, you will see a message telling you
so
[nsid@platolgn01 ~]$ module list
Currently Loaded Modules:
1) CCconfig 4) imkl/2020.1.217 (math) 7) libfabric/1.10.1
2) gentoo/2020 (S) 5) intel/2020.1.217 (t) 8) openmpi/4.0.3 (m)
3) gcccore/.9.3.0 (H) 6) ucx/1.8.0 9) StdEnv/2020 (S)
[Some output removed for clarity]
Loading and unloading software
To load a software module, use module load.
In this example we will use R, a software environment for statistics.
Initially, R is not loaded.
We can test this by using the which command.
which looks for programs the same way that Bash does,
so we can use it to tell us where a particular piece of software is stored.
[nsid@platolgn01 ~]$ which R
/usr/bin/which: no R in (/cvmfs/soft.computecanada.ca/easybuild/software/2020/avx2/Compiler/intel2020/openmpi/4.0.3/bin:/cvmfs/soft.computecanada.ca/easybuild/software/2020/avx2/Core/libfabric/1.10.1/bin:/cvmfs/soft.computecanada.ca/easybuild/software/2020/avx2/Core/ucx/1.8.0/bin:/cvmfs/restricted.computecanada.ca/easybuild/software/2020/Core/intel/2020.1.217/compilers_and_libraries_2020.1.217/linux/bin/intel64:/cvmfs/soft.computecanada.ca/easybuild/software/2020/Core/gcccore/9.3.0/bin:/cvmfs/soft.computecanada.ca/easybuild/bin:/cvmfs/soft.computecanada.ca/custom/bin:/cvmfs/soft.computecanada.ca/gentoo/2020/usr/sbin:/cvmfs/soft.computecanada.ca/gentoo/2020/usr/bin:/cvmfs/soft.computecanada.ca/gentoo/2020/sbin:/cvmfs/soft.computecanada.ca/gentoo/2020/bin:/cvmfs/soft.computecanada.ca/custom/bin/computecanada:/globalhome/olf067/HPC/.local/bin:/cm/shared/bin:/opt/software/bin:/opt/software/slurm/bin:/usr/local/bin:/usr/bin:/usr/local/sbin:/usr/sbin:/opt/dell/srvadmin/bin)
We can load the R command with module load (be careful of the case of the
letters, r vs R):
[nsid@platolgn01 ~]$ module load r
[nsid@platolgn01 ~]$ which R
/cvmfs/soft.computecanada.ca/easybuild/software/2020/avx2/Core/r/4.3.1/bin/R
So, what just happened?
To understand the output, first we need to understand the nature of the $PATH environment
variable. $PATH is a special environment variable that controls where a UNIX system looks for
software. Specifically $PATH is a list of directories (separated by :) that the OS searches
through for a command before giving up and telling us it can’t find it. As with all environment
variables we can print it out using echo.
[nsid@platolgn01 ~]$ echo $PATH
/cvmfs/soft.computecanada.ca/easybuild/software/2020/avx2/Core/r/4.3.1/bin:/cvmfs/soft.computecanada.ca/easybuild/software/2020/Core/java/13.0.2:/cvmfs/soft.computecanada.ca/easybuild/software/2020/Core/java/13.0.2/bin:/cvmfs/soft.computecanada.ca/easybuild/software/2020/Core/flexiblas/3.0.4/bin:/cvmfs/soft.computecanada.ca/easybuild/software/2020/avx2/Compiler/intel2020/openmpi/4.0.3/bin:/cvmfs/soft.computecanada.ca/easybuild/software/2020/avx2/Core/libfabric/1.10.1/bin:/cvmfs/soft.computecanada.ca/easybuild/software/2020/avx2/Core/ucx/1.8.0/bin:/cvmfs/restricted.computecanada.ca/easybuild/software/2020/Core/intel/2020.1.217/compilers_and_libraries_2020.1.217/linux/bin/intel64:/cvmfs/soft.computecanada.ca/easybuild/software/2020/Core/gcccore/9.3.0/bin:/cvmfs/soft.computecanada.ca/easybuild/bin:/cvmfs/soft.computecanada.ca/custom/bin:/cvmfs/soft.computecanada.ca/gentoo/2020/usr/sbin:/cvmfs/soft.computecanada.ca/gentoo/2020/usr/bin:/cvmfs/soft.computecanada.ca/gentoo/2020/sbin:/cvmfs/soft.computecanada.ca/gentoo/2020/bin:/cvmfs/soft.computecanada.ca/custom/bin/computecanada:/globalhome/olf067/HPC/.local/bin:/cm/shared/bin:/opt/software/bin:/opt/software/slurm/bin:/usr/local/bin:/usr/bin:/usr/local/sbin:/usr/sbin:/opt/dell/srvadmin/bin
You’ll notice a similarity to the output of the which command. In this case, there’s only one
difference: the different directory at the beginning. When we ran the module load command,
it added a directory to the beginning of our $PATH. Let’s examine what’s there:
[nsid@platolgn01 ~]$ ls /cvmfs/soft.computecanada.ca/easybuild/software/2020/avx2/Core/r/4.3.1/bin
R* Rscript*
Taking this to its conclusion, module load will add software to your $PATH. It “loads”
software. The module load command will also load required software dependencies.
To demonstrate, let’s use module list. module list shows all loaded software modules.
[nsid@platolgn01 ~]$ module list
Currently Loaded Modules:
1) CCconfig 5) intel/2020.1.217 (t) 9) StdEnv/2020 (S)
2) gentoo/2020 (S) 6) ucx/1.8.0 10) flexiblascore/.3.0.4 (H)
3) gcccore/.9.3.0 (H) 7) libfabric/1.10.1 11) java/13.0.2 (t)
4) imkl/2020.1.217 (math) 8) openmpi/4.0.3 (m) 12) r/4.3.1 (t)
[Some output removed for clarity]
In this case, loading the r module also loaded java/13.0.2.
[nsid@platolgn01 ~]$ module unload r
[nsid@platolgn01 ~]$ module list
Currently Loaded Modules:
1) CCconfig 4) imkl/2020.1.217 (math) 7) libfabric/1.10.1
2) gentoo/2020 (S) 5) intel/2020.1.217 (t) 8) openmpi/4.0.3 (m)
3) gcccore/.9.3.0 (H) 6) ucx/1.8.0 9) StdEnv/2020 (S)
[Some output removed for clarity]
So using module unload “un-loads” a module along with its dependencies. If we wanted to unload
everything at once, we could run module purge.
[nsid@platolgn01 ~]$ module load r
[nsid@platolgn01 ~]$ module load python
[nsid@platolgn01 ~]$ module purge
The following modules were not unloaded:
(Use "module --force purge" to unload all):
1) CCconfig 4) imkl/2020.1.217 7) libfabric/1.10.1
2) gentoo/2020 5) intel/2020.1.217 8) openmpi/4.0.3
3) gcccore/.9.3.0 6) ucx/1.8.0 9) StdEnv/2020
Note that module purge does not, by default, unload the core modules that were
already loaded when we logged in. Therefore, module purge is useful to “reset”
your environment.
Software versioning
So far, we’ve learned how to load and unload software packages. This is very useful. However, we have not yet addressed the issue of software versioning. At some point or other, you will run into issues where only one particular version of some software will be suitable. Perhaps a key bugfix only happened in a certain version, or version X broke compatibility with a file format you use. In either of these example cases, it helps to be very specific about what software is loaded.
Let’s examine the output of module avail gcc.
[nsid@platolgn01 ~]$ module avail gcc
[Some output removed for clarity]
-------------------------------------- Core Modules --------------------------------------
gcc/8.4.0 (t) gcc/10.2.0 (t) gcc/11.3.0 (t)
gcc/9.3.0 (t,D) gcc/10.3.0 (t)
[Some output removed for clarity]
Let’s take a closer look at the gcc module. GCC is an extremely widely used C/C++/Fortran
compiler. Tons of software is dependent on the GCC version, and might not compile or run if the
wrong version is loaded. In this case, several versions are available.
How do we load each one and which one is the default?
In this case, gcc/9.3.0 has a (D) next to it. This indicates that it is the default - if we type
module load gcc, this is the version that will be loaded.
[nsid@platolgn01 ~]$ module load gcc
[nsid@platolgn01 ~]$ gcc --version
Lmod is automatically replacing "intel/2020.1.217" with "gcc/9.3.0".
Due to MODULEPATH changes, the following have been reloaded:
1) openmpi/4.0.3
gcc (GCC) 9.3.0
Copyright (C) 2019 Free Software Foundation, Inc.
This is free software; see the source for copying conditions. There is NO
warranty; not even for MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE.
Note that three things happened: the default version of GCC was loaded (version 9.3.0), the Intel
compilers (which conflict with GCC) were unloaded, and software that is dependent on compiler
(OpenMPI) was reloaded. The module system turned what might be a super-complex operation into a
single command.
So how do we load a non-default version of a software package? In this case, the
only change we need to make is be more specific about the module we are loading
by leaving in the version number after the /.
[nsid@platolgn01 ~]$ module load gcc/10.2.0
[nsid@platolgn01 ~]$ gcc --version
Inactive Modules:
1) openmpi
The following have been reloaded with a version change:
1) gcc/9.3.0 => gcc/10.2.0 2) gcccore/.9.3.0 => gcccore/.10.2.0
gcc (GCC) 10.2.0
Copyright (C) 2020 Free Software Foundation, Inc.
This is free software; see the source for copying conditions. There is NO
warranty; not even for MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE.
We now have successfully switched from GCC 9.3.0 to GCC 10.2.0. Because OpenMPI is not available for that specific version of GCC, its module was inactivated.
Using software modules in scripts
Create a job that is able to run
R --version. Remember, no software is loaded by default! Running a job is just like logging on to the system (you should not assume a module loaded on the login node is loaded on a compute node).Solution
[nsid@platolgn01 ~]$ nano job-with-module.sh [nsid@platolgn01 ~]$ cat job-with-module.sh#!/bin/bash module load r/4.3.1 R --version[nsid@platolgn01 ~]$ sbatch job-with-module.sh
Key Points
Load software with
module load softwareNameUnload software with
module purgeThe module system handles software versioning and package conflicts for you automatically.
Transferring files
Overview
Teaching: 30 min
Exercises: 10 minQuestions
How do I upload/download files to the cluster?
Objectives
Be able to transfer files to and from a computing cluster.
Computing with a remote computer offers very limited use if we cannot get files to or from the cluster. There are several options for transferring data between computing resources.
Download files from the internet using wget
One of the most straightforward ways to download files is to use wget. Any file that can be
downloaded in your web browser with an accessible link can be downloaded using wget. This is a
quick way to download datasets or source code.
The syntax is: wget https://some/link/to/a/file.tar.gz. For example, download the lesson sample
files using the following command:
[nsid@platolgn01 ~]$ wget https://ofisette.github.io/hpc-intro-plato/files/bash-lesson.tar.gz
Transferring single files and folders with scp
To copy a single file to or from the cluster, we can use scp (“secure copy”). The syntax can be
a little complex for new users, but we’ll break it down.
To transfer to another computer:
[user@laptop ~]$ scp path/to/local/file.txt nsid@plato.usask.ca:path/on/Plato
Transfer a file
Create a “calling card” with your name and email address, then transfer it to your home directory on Plato.
Solution
Create a file like this, with your name (or an alias) and top-level domain:
[user@laptop ~]$ cat calling-card.txtYour Name Your.Address@institution.tldNow, transfer it to Plato:
[user@laptop ~]$ scp calling-card.txt nsid@plato.usask.ca:/globalhome/nsid/HPC/calling-card.txt 100% 37 7.6 KB/s 00:00
We can often simplify the path given to the command. On the remote computer,
everything after the : is relative to our home directory. A single : would
put a file directly in your home directory.
[user@laptop ~]$ scp local-file.txt nsid@plato.usask.ca:
To recursively copy a directory, we just add the -r (recursive) flag:
[user@laptop ~]$ scp -r some-local-folder/ nsid@plato.usask.ca:
This will create the directory some-local-folder on the remote system,
and recursively copy all the content from the local to the remote system.
Existing files on the remote system will not be modified, unless
there are files from the local system with the same name, in which
case the remote files will be overwritten.
The trailing slashes in the directory names are optional, and have no effect for scp
-r, but they are important in other commands, like rsync.
To download from another computer:
[user@laptop ~]$ scp nsid@plato.usask.ca:path/on/Plato/file.txt path/to/local/
A note on
rsyncAs you gain experience with transferring files, you may find the
scpcommand limiting. The rsync utility provides advanced features for file transfer and is typically faster compared to bothscpandsftp(see below). It is especially useful for transferring large and/or many files and creating synced backup folders.The syntax is similar to
scp. To transfer to another computer with commonly used options:[user@laptop ~]$ rsync -rvzP path/to/local/file.txt nsid@plato.usask.ca:directory/path/on/Plato/The
r(recursive) option copies files and directories recursively; thev(verbose) option gives verbose output to help monitor the transfer; thez(compression) option compresses the file during transit to reduce size and transfer time; and theP(partial/progress) option preserves partially transferred files in case of an interruption and also displays the progress of the transfer.To recursively copy a directory, we can use the same options:
[user@laptop ~]$ rsync -rvzP path/to/local/dir nsid@plato.usask.ca:directory/path/on/Plato/As written, this will place the local directory and its contents under the specified directory on the remote system. If the trailing slash is omitted on the destination, a new directory corresponding to the transferred directory (‘dir’ in the example) will not be created, and the contents of the source directory will be copied directly into the destination directory.
To download a file, we simply change the source and destination:
[user@laptop ~]$ rsync -rvzP nsid@plato.usask.ca:path/on/Plato/file.txt path/to/local/
Archiving files
One of the biggest challenges we often face when transferring data between remote HPC systems is that of large numbers of files. There is an overhead to transferring each individual file and when we are transferring large numbers of files these overheads combine to slow down our transfers to a large degree.
The solution to this problem is to archive multiple files into smaller numbers of larger files before we transfer the data to improve our transfer efficiency. Sometimes we will combine archiving with compression to reduce the amount of data we have to transfer and so speed up the transfer.
The most common archiving command you will use on a (Linux) HPC cluster is tar. tar can be used
to combine files into a single archive file and, optionally, compress. For example, to collect
all files contained inside output_data into an archive file called output_data.tar we would use:
[user@laptop ~]$ tar -cvf output_data.tar output_data/
The options we used for tar are:
-c- Create new archive-v- Verbose (print what you are doing!)-f mydata.tar- Create the archive in file output_data.tar
The tar command allows users to concatenate flags. Instead of typing tar -c -v -f, we can use
tar -cvf.
The tar command can also be used to interrogate and unpack archive files. The -t argument
(“table of contents”) lists the contents of the referred-to file without unpacking it.
The -x (“extract”) flag unpacks the referred-to file. To unpack the file after we have
transferred it:
[user@laptop ~]$ tar -xvf output_data.tar
This will put the data into a directory called output_data. Be careful, it will overwrite data
there if this directory already exists!
Sometimes you may also want to compress the archive to save space and speed up the transfer.
However, you should be aware that for large amounts of data compressing and un-compressing can take
longer than transferring the un-compressed data so you may not want to transfer. To create a
compressed archive using tar we add the -z option and add the .gz extension to the file to
indicate it is gzip-compressed, e.g.:
[user@laptop ~]$ tar -czvf output_data.tar.gz output_data/
The tar command is used to extract the files from the archive in exactly the same way as for
uncompressed data. The tar command recognizes that the data is compressed, and automatically
selects the correct decompression algorithm at the time of extraction:
[user@laptop ~]$ tar -xvf output_data.tar.gz
Transferring files
Using one of the above methods, try transferring files to and from the cluster. Which method do you like the best?
Working with Windows
When you transfer files to from a Windows system to a Unix system (Mac, Linux, BSD, Solaris, etc.) this can cause problems. Windows encodes its files slightly different than Unix, and adds an extra character to every line.
On a Unix system, every line in a file ends with a
\n(newline). On Windows, every line in a file ends with a\r\n(carriage return + newline). This causes problems sometimes.Though most modern programming languages and software handles this correctly, in some rare instances, you may run into an issue. The solution is to convert a file from Windows to Unix encoding with the
dos2unixcommand.You can identify if a file has Windows line endings with
cat -A filename. A file with Windows line endings will have^M$at the end of every line. A file with Unix line endings will have$at the end of a line.To convert the file, just run
dos2unix filename. (Conversely, to convert back to Windows format, you can rununix2dos filename.)
A note on ports
All file transfers using the above methods use encrypted communication over port 22. This is the same connection method used by SSH. In fact, all file transfers using these methods occur through an SSH connection. If you can connect via SSH over the normal port, you will be able to transfer files.
Key Points
wgetdownloads a file from the internet.
scptransfer files to and from your computer.
Running a parallel job
Overview
Teaching: 10 min
Exercises: 5 minQuestions
How do we execute a task in parallel?
Objectives
Understand how to run a parallel job on a cluster.
We now have the tools we need to run a multi-processor job. This is a very important aspect of HPC systems, as parallelism is one of the primary tools we have to improve the performance of computationnal tasks.
Running the Parallel Job
We will run an example that uses the Message Passing Interface (MPI) for parallelism – this is a common tool on HPC systems.
What is MPI?
The Message Passing Interface is a set of tools which allow multiple parallel jobs to communicate with each other. Typically, a single executable is run multiple times, possibly on different machines, and the MPI tools are used to inform each instance of the executable about how many instances there are, which instance it is. MPI also provides communication tools to allow communication and coordination between instances. An MPI instance typically has its own copy of all the local variables.
MPI jobs cannot generally be run as stand-alone executables.
Instead, they should be started with the mpirun command, which
ensures that the appropriate run-time support for parallelism
is included.
On its own, mpirun can take many arguments
specifying how many machines will partcipate in the process.
In the context of our queuing system, however, we do not need
to specify this information, the mpirun command will obtain
it from the queuing system, by examining the environment
variables set when the job is launched.
Our example implements a stochastic algorithm for estimating the value of pi, the ratio of the circumference to the diameter of a circle. The program generates a large number of random points on a 2x2 square centered on the origin, and checks how many of these points fall inside the unit circle. On average, pi/4 of the randomly-selected points should fall in the circle, so pi can be estimated from 4f, where f is the observed fraction of points that fall in the circle. Because each sample is independent, this algorithm is easily implemented in parallel.
We have provided a Python implementation, which uses MPI and Numpy.
Download the Python executable and make it executable using the following commands:
[nsid@platolgn01 ~]$ wget https://ofisette.github.io/hpc-intro-plato/files/pi.py
[nsid@platolgn01 ~]$ chmod +x pi.py
Feel free to examine the code in an editor – it is richly commented, and should provide some educational value.
Our purpose here is to exercise the parallel workflow of the cluster.
Create a submission file, requesting more than one task on a single node:
[nsid@platolgn01 ~]$ nano parallel-example.sh
[nsid@platolgn01 ~]$ cat parallel-example.sh
#!/bin/bash
#SBATCH --job-name=pi-montecarlo
#SBATCH --ntasks=4
module load gcc/9.3.0
module load python/3.8
module load scipy-stack/2020a
module load mpi4py
mpirun ./pi.py
Then submit your job.
[nsid@platolgn01 ~]$ sbatch parallel-example.sh
As before, use the status commands to check when your job runs,
and use ls to locate the output file, then examine it.
Is it what you expected? How good is the value for pi?
Key Points
Parallelism is an important feature of HPC clusters.
MPI parallelism is a common case.
The queuing system facilitates executing parallel tasks.
Using resources effectively
Overview
Teaching: 15 min
Exercises: 10 minQuestions
How do we monitor our jobs?
How can I get my jobs scheduled more easily?
Objectives
Understand how to look up job statistics and profile code.
Understand job size implications.
We now know virtually everything we need to know about getting stuff on a cluster. We can log on, submit different types of jobs, use pre-installed software, and install and use software of our own. What we need to do now is use the systems effectively.
Estimating required resources using the scheduler
Although we covered requesting resources from the scheduler earlier, how do we know what type of resources the software will need in the first place, and the extent of its demand for each?
Unless the developers or prior users have provided some idea, we don’t. Not until we’ve tried it ourselves at least once. We’ll need to benchmark our job and experiment with it before we know how how great its demand for system resources.
Read the documentation
Most HPC facilities maintain documentation as a wiki, website, or a document sent along when you register for an account. Take a look at these resources, and search for the software of interest: somebody might have written up guidance for getting the most out of it.
The most effective way of figuring out the resources required for a job to run successfully needs is
to submit a test job, and then ask the scheduler about its impact using sacct.
[nsid@platolgn01 ~]$ sacct
This shows all the jobs we ran recently (note that there are multiple entries per job). To get info about a specific job, we change command slightly.
[nsid@platolgn01 ~]$ sacct -j 727107
This shows a lot of information, even more if you use the long display option,
-l. Plato also has a convenience command, seff, that gives a terser output.
[nsid@platolgn01 ~]$ seff 727107
Job ID: 727107
Cluster: plato
User/Group: nsid/nsid
State: COMPLETED (exit code 0)
Nodes: 1
Cores per node: 4
CPU Utilized: 00:00:04
CPU Efficiency: 25.00% of 00:00:16 core-walltime
Job Wall-clock time: 00:00:04
Memory Utilized: 1.12 MB
Memory Efficiency: 0.05% of 2.00 GB
Some interesting fields include the following:
- Hostname: Where did your job run?
- MaxRSS: What was the maximum amount of memory used?
- Elapsed: How long did the job take?
- State: What is the job currently doing/what happened to it?
- MaxDiskRead: Amount of data read from disk.
- MaxDiskWrite: Amount of data written to disk.
You can use this knowledge to set up the next job with a close estimate of its load on the system. A good general rule is to ask the scheduler for 20% to 30% more time and memory than you expect the job to need. This ensures that minor fluctuations in run time or memory use will not result in your job being cancelled by the scheduler. Keep in mind that if you ask for too much, your job may not run even though enough resources are available, because the scheduler will be waiting to match what you asked for.
Measuring the statistics of currently running tasks
Connecting to Nodes
Typically, clusters allow users to connect to compute nodes where they have
running jobs. This is useful to check on a running job and see how it’s doing.
To know which node to connect to, use squeue. Then, run ssh
nodename. Once you are on the node of interest, use programs such as top or
ps, as described below.
Monitor system processes with top
The most reliable way to check current system stats is with top. Some sample output might look
like the following (type q to exit top):
[nsid@platolgn01 ~]$ top
top - 21:00:19 up 3:07, 1 user, load average: 1.06, 1.05, 0.96
Tasks: 311 total, 1 running, 222 sleeping, 0 stopped, 0 zombie
%Cpu(s): 7.2 us, 3.2 sy, 0.0 ni, 89.0 id, 0.0 wa, 0.2 hi, 0.2 si, 0.0 st
KiB Mem : 16303428 total, 8454704 free, 3194668 used, 4654056 buff/cache
KiB Swap: 8220668 total, 8220668 free, 0 used. 11628168 avail Mem
PID USER PR NI VIRT RES SHR S %CPU %MEM TIME+ COMMAND
1693 jeff 20 0 4270580 346944 171372 S 29.8 2.1 9:31.89 gnome-shell
3140 jeff 20 0 3142044 928972 389716 S 27.5 5.7 13:30.29 Web Content
3057 jeff 20 0 3115900 521368 231288 S 18.9 3.2 10:27.71 firefox
6007 jeff 20 0 813992 112336 75592 S 4.3 0.7 0:28.25 tilix
1742 jeff 20 0 975080 164508 130624 S 2.0 1.0 3:29.83 Xwayland
1 root 20 0 230484 11924 7544 S 0.3 0.1 0:06.08 systemd
68 root 20 0 0 0 0 I 0.3 0.0 0:01.25 kworker/4:1
2913 jeff 20 0 965620 47892 37432 S 0.3 0.3 0:11.76 code
2 root 20 0 0 0 0 S 0.0 0.0 0:00.02 kthreadd
Overview of the most important fields:
PID: What is the numerical id of each process?USER: Who started the process?RES: What is the amount of memory currently being used by a process (in bytes)?%CPU: How much of a CPU is each process using? Values higher than 100 percent indicate that a process is running in parallel.%MEM: What percent of system memory is a process using?TIME+: How much CPU time has a process used so far? Processes using 2 CPUs accumulate time at twice the normal rate.COMMAND: What command was used to launch a process?
htop provides a curses-based overlay for top, producing a better-organized and “prettier”
dashboard in your terminal.
ps
To show all processes from your current session, type ps.
[nsid@platolgn01 ~]$ ps
PID TTY TIME CMD
15113 pts/5 00:00:00 bash
15218 pts/5 00:00:00 ps
Note that this will only show processes from our current session. To show all processes you own
(regardless of whether they are part of your current session or not), you can use ps ux.
[nsid@platolgn01 ~]$ ps ux
USER PID %CPU %MEM VSZ RSS TTY STAT START TIME COMMAND
nsid 67780 0.0 0.0 149140 1724 pts/81 R+ 13:51 0:00 ps ux
nsid 73083 0.0 0.0 142392 2136 ? S 12:50 0:00 sshd: nsid@pts/81
nsid 73087 0.0 0.0 114636 3312 pts/81 Ss 12:50 0:00 -bash
This is useful for identifying which processes are doing what.
Key Points
The smaller your job, the faster it will schedule.
Using shared resources responsibly
Overview
Teaching: 15 min
Exercises: 5 minQuestions
How can I be a responsible user?
How can I protect my data?
How can I best get large amounts of data off an HPC system?
Objectives
Learn how to be a considerate shared system citizen.
Understand how to protect your critical data.
Appreciate the challenges with transferring large amounts of data off HPC systems.
Understand how to convert many files to a single archive file using tar.
One of the major differences between using remote HPC resources and your own system (e.g. your laptop) is that remote resources are shared. How many users the resource is shared between at any one time varies from system to system but it is unlikely you will ever be the only user logged into or using such a system.
The widespread usage of scheduling systems where users submit jobs on HPC resources is a natural outcome of the shared nature of these resources. There are other things you, as an upstanding member of the community, need to consider.
Be kind to the login nodes
The login node is often busy managing all of the logged in users, creating and editing files and compiling software. If the machine runs out of memory or processing capacity, it will become very slow and unusable for everyone. While the machine is meant to be used, be sure to do so responsibly, in ways that will not adversely impact other users’ experience.
Login nodes are always the right place to launch jobs. Cluster policies vary, but they may also be used for proving out workflows, and in some cases, may host advanced cluster-specific debugging or development tools. The cluster may have modules that need to be loaded, possibly in a certain order, and paths or library versions that differ from your laptop, and doing an interactive test run on the head node is a quick and reliable way to discover and fix these issues.
Login nodes are a shared resource
Remember, the login node is shared with all other users and your actions could cause issues for other people. Think carefully about the potential implications of issuing commands that may use large amounts of resource.
Unsure? Ask your friendly systems administrator (“sysadmin”) if the thing you’re contemplating is suitable for the login node, or if there’s another mechanism to get it done safely.
You can always use the commands top and ps ux to list the processes that are
running on the login node along with the amount of CPU and memory they are
using. If this check reveals that the login node is somewhat idle, you can
safely use it for your non-routine processing task. If something goes wrong,
e.g. the process takes too long or doesn’t respond, you can use the kill
command along with the PID to terminate the process.
Login Node Etiquette
Which of these commands would be a routine task to run on the login node?
python physics_sim.pymakecreate_directories.shmolecular_dynamics_2tar -xzf R-3.3.0.tar.gzSolution
Building software, creating directories, and unpacking software are common and acceptable tasks for the login node: options #2 (
make), #3 (mkdir), and #5 (tar) are probably OK. Note that script names do not always reflect their contents: before launching #3, pleaseless create_directories.shand make sure it’s not a Trojan horse.Running resource-intensive applications is frowned upon. Unless you are sure it will not affect other users, do not run jobs like #1 (
python) or #4 (custom MD code).
Test before scaling
Remember that you are generally charged for usage on shared systems. A simple mistake in a job script can end up costing a large amount of resource budget. Imagine a job script with a mistake that makes it sit doing nothing for 24 hours on 1000 cores or one where you have requested 2000 cores by mistake and only use 100 of them! This problem can be compounded when people write scripts that automate job submission (for example, when running the same calculation or analysis over lots of different parameters or files). When this happens it hurts both you (as you waste lots of charged resource) and other users (who are blocked from accessing the idle compute nodes).
On very busy resources you may wait many days in a queue for your job to fail within 10 seconds of starting due to a trivial typo in the job script. This is extremely frustrating! Most systems provide dedicated resources for testing that have short wait times to help you avoid this issue.
Test job submission scripts that use large amounts of resources
Before submitting a large run of jobs, submit one as a test first to make sure everything works as expected.
Before submitting a very large or very long job submit a short truncated test to ensure that the job starts as expected.
Have a backup plan
Although many HPC systems keep backups, it does not always cover all the file systems available and may only be for disaster recovery purposes (i.e. for restoring the whole file system if lost rather than an individual file or directory you have deleted by mistake). Protecting critical data from corruption or deletion is primarily your responsibility: keep your own backup copies.
Version control systems (such as Git) often have free, cloud-based offerings (e.g., GitHub and GitLab) that are generally used for storing source code. Even if you are not writing your own programs, these can be very useful for storing job scripts, analysis scripts and small input files.
For larger amounts of data, you should make sure you have a robust system in place for taking
copies of critical data off the HPC system wherever possible to backed-up storage. Tools such
as rsync can be very useful for this.
Your access to the shared HPC system will generally be time-limited so you should ensure you have a plan for transferring your data off the system before your access finishes. The time required to transfer large amounts of data should not be underestimated and you should ensure you have planned for this early enough (ideally, before you even start using the system for your research).
In all these cases, the helpdesk of the system you are using should be able to provide useful guidance on your options for data transfer for the volumes of data you will be using.
Your data is your responsibility
Make sure you understand what the backup policy is on the file systems on the system you are using and what implications this has for your work if you lose your data on the system. Plan your backups of critical data and how you will transfer data off the system throughout the project.
Transferring data
As mentioned above, many users run into the challenge of transferring large amounts of data off HPC systems at some point (this is more often in transferring data off than onto systems but the advice below applies in either case). Data transfer speed may be limited by many different factors so the best data transfer mechanism to use depends on the type of data being transferred and where the data is going. Some of the key issues to be aware of are:
- Disk speed: File systems on HPC systems are often highly parallel, consisting of a very large number of high performance disk drives. This allows them to support a very high data bandwidth. Unless the remote system has a similar parallel file system you may find your transfer speed limited by disk performance at that end.
- Meta-data performance: Meta-data operations such as opening and closing files or listing the owner or size of a file are much less parallel than read/write operations. If your data consists of a very large number of small files you may find your transfer speed is limited by meta-data operations. Meta-data operations performed by other users of the system can also interact strongly with those you perform so reducing the number of such operations you use (by combining multiple files into a single file) may reduce variability in your transfer rates and increase transfer speeds.
- Network speed: Data transfer performance can be limited by network speed. More importantly it is limited by the slowest section of the network between source and destination. If you are transferring to your laptop/workstation, this is likely to be its connection (either via LAN or wifi).
- Firewall speed: Most modern networks are protected by some form of firewall that filters out malicious traffic. This filtering has some overhead and can result in a reduction in data transfer performance. The needs of a general purpose network that hosts email/web-servers and desktop machines are quite different from a research network that needs to support high volume data transfers. If you are trying to transfer data to or from a host on a general purpose network you may find the firewall for that network will limit the transfer rate you can achieve.
As mentioned above, if you have related data that consists of a large number of small files it
is strongly recommended to pack the files into a larger archive file for long term storage and
transfer. A single large file makes more efficient use of the file system and is easier to move,
copy and transfer because significantly fewer metadata operations are required. Archive files can
be created using tools like tar and zip. We have already met tar when we talked about data
transfer earlier.
Consider the best way to transfer data
If you are transferring large amounts of data you will need to think about what may affect your transfer performance. It is always useful to run some tests that you can use to extrapolate how long it will take to transfer your data.
Say you have a “data” folder containing 10,000 or so files, a healthy mix of small and large ASCII and binary data. Which of the following would be the best way to transfer them to Plato?
[user@laptop ~]$ scp -r data nsid@plato.usask.ca:~/[user@laptop ~]$ rsync -rz data nsid@plato.usask.ca:~/[user@laptop ~]$ tar -cvf data.tar data [user@laptop ~]$ rsync -rz data.tar nsid@plato.usask.ca:~/[user@laptop ~]$ tar -cvzf data.tar.gz data [user@laptop ~]$ rsync -r data.tar.gz nsid@plato.usask.ca:~/Solution
scpwill recursively copy the directory. This works, but without compression.rsync -rzadds compression, which will save some bandwidth. If you have a strong CPU at both ends of the line, and you’re on a slow network, this is a good choice.- This command first uses
tarto merge everything into a single file, thenrsync -zto transfer it with compression. With this large number of files, metadata overhead can hamper your transfer, so this is a good idea.- This command uses
tar -zto compress the archive, thenrsyncto transfer it. This may perform similarly to #3, but in most cases (for large datasets), it’s the best combination of high throughput and low latency (making the most of your time and network connection).
Key Points
Be careful how you use the login node.
Your data on the system is your responsibility.
Plan and test large data transfers.
It is often best to convert many files to a single archive file before transferring.
Again, don’t run stuff on the login node.